eDNA: Making the invisible visible ¶
Every living organism leaves traces of its genetic material in the environment, for example in the form of skin flakes, pollen, hair, or droppings. We analyze this environmental DNA, or eDNA for short, to detect animals, plants, fungi, and microorganisms.
eDNA-based methods are revolutionizing biodiversity monitoring in many ways:
- Non-invasive sampling: A water, soil, or air sample is enought – with minimal disturbance to the organisms themselves.
- High sensitivity: Even highly fragmented or tiny amounts of DNA can be reliably analyzed.
- Huge amount of information: Just one cubic centimeter of an environmental sample can yield gigabytes of DNA sequence data.
- High efficiency: Even rare or elusive species can be detected.
Despite the many advantages and cost-effectiveness of eDNA approaches, classical monitoring methods remain indispensable – not least because they are based on the knowledge of experienced specialists.
How we analyze eDNA ¶
An eDNA analysis requires the utmost care to draw reliable and complex conclusions:
- We exclusively use DNA-free material for sampling.
- eDNA samples often contain only tiny amounts of DNA fragments. In such cases, we work in the special clean-air laboratory at WSL to avoid contamination.
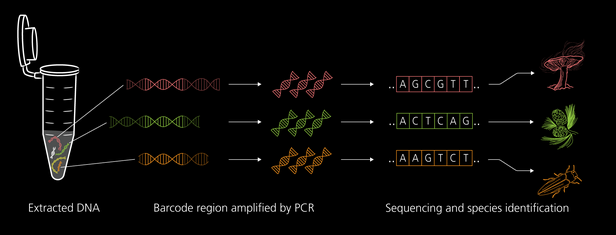
- Metabarcoding allows us to detect many species in a sample at the same time. We amplify a specific DNA segment using PCR to obtain enough material for analysis. This segment, also known as the barcode region, is selected so that it is present in the genome of all species within a target group.
- Using powerful bioinformatics programs, we analyze the composition of the samples and match the sequences against reference databases. This allows us to identify the species or at least higher taxonomic groups in the environmental sample.
What we use eDNA analyses for ¶

Findings from eDNA analyses have a wide range of applications in environmental research and conservation. Here are a few examples from projects at WSL:
- We map the distribution of fungal species in Switzerland for the Red List of threatened species.
- We detect invasive species such as the emerald ash borer at an early stage.
- We compare species communities across different habitats.
- We identify which amphibians occur in ponds and which wild animals live in the city of Zurich.
- We conduct large-scale studies of soil biodiversity in Swiss forest soils.
eDNA methods hold great potential for further development, and we at WSL are actively contributing to this progress. Thanks to even more precise technologies, we will also be able to reanalyze older, frozen DNA samples in the future.
Data from eDNA-based projects at WSL, ETH Zurich, and the global WildinSync initiative are available on WSL’s data portal EnviDAT.